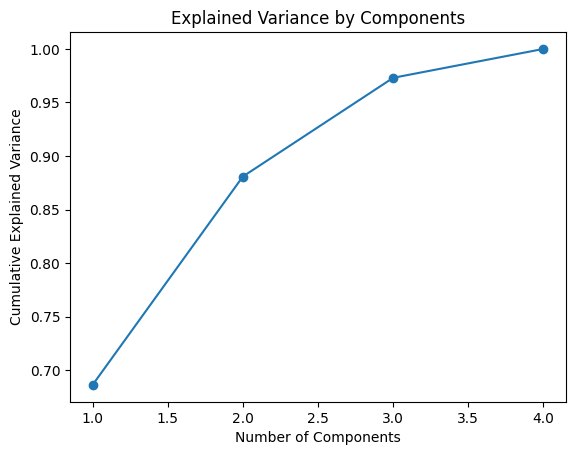
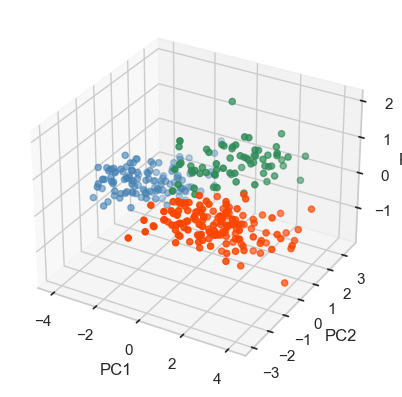
import holopheno
import pandas as pd
from palmerpenguins import load_penguins
penguins = load_penguins()
x_columns = ['species', 'island', 'sex']
y_columns = ['bill_length_mm',
'bill_depth_mm',
'flipper_length_mm',
'body_mass_g', ]
penguins_h = holopheno.read_data(penguins, x = x_columns, y = y_columns);Data info:
sample_size 333
unique species values ['Adelie' 'Gentoo' 'Chinstrap']
unique island values ['Torgersen' 'Biscoe' 'Dream']
unique sex values ['male' 'female']